Clinical Analysis of OncomiR - Therapeutic Targeting of Tumorigenesis and Tumor Disease
Biaoru Li
Georgia Cancer Center, MCG, Augusta, GA, USA.
*Corresponding Author
Biaoru Li,
Associated Professor,
Georgia Cancer Center, MCG,
Augusta, GA, 30912, USA.
E-mail: BLI@gru.edu
Received: February 17, 2017; Accepted: March 01, 2017; Published: March 02, 2017
Citation: Biaoru Li (2017) Clinical Analysis of OncomiR -Therapeutic Targeting of Tumorigenesis and Tumor Disease. Int J Genomics Proteomics Metabolomics Bioinformatics. 2(2), 28-35.
Copyright: Biaoru Li© 2017. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
A microRNA alteration discovered in clinical patients has largely studied in tumor diseases. Now many profiles of microRNAs alterations have been uncovered to associate with tumor disease and tumorigenesis, and thus the profiles of the miRNA alteration are called as “OncomiR”. Clinically, detecting miRNA profiles from patients has been increasingly applied to predict prognosis and monitor clinical response to treat tumor diseases, classify tumor diseases and study utilizing miRNA profiles to perform RNA therapy for tumor disease and tumorigenesis. The manual aims focus on patient diagnosis, treatment and prognosis prediction by the oncomiR performance and analysis. In order to make clinical physicians easy to understand a practicable procedure for the oncomiR application, the contents include clinical sampling for miRNA performance, clinical miRNA detection with their analyses and clinical employment including prediction of prognosis of tumor disease and prediction of response to chemotherapy, diagnosis of tumor classification from miRNA biomarkers and RNA therapy from the miRNA analysis. In final part, I will present challenges and future development based on the clinical “oncomiR”.
2.Introduction
3.Clinical Sampling
3.1 Non-tumor tissues sampling
3.2 Tumor cell sampling from tumor tissue
4.Clinical MiRNA Detection with their Analyses
4.1 MiRNA qPCR assays
4.2 Multiplex miRNA profiling
4.3 MiRNA Arrays
4.4 RNA-seq
4.5 MiRNA Target Prediction Tool for Further Analysis
5.Clinical Application
5.1 Prediction of Prognosis/Drug Responses to Chemotherapy
5.2 Diagnosis for Tumor Classification and Biomarkers
5.3 MiRNA Treatment and Clinical Trials
6.Conclusion
6.1 Clinical oncomiR Diagnoses Require Routine Specificity and Sensitivity Test
6.2 Clinical oncomiR Diagnoses Require Confirmation Assay
6.3 OncomiR by Quantitative Network Support
7.Acknowledgments
8.References
KeyWords
Clinical Sampling; cfmiRNA; Exosome; OncomiR (oncomiRs); miRNA; siRNA; Tumor Prediction; Tumor Prognosis and Biomarkers.
Introduction
A microRNA (miRNA) with 22 nucleotides non-coding RNA exists in the intracellular and extracellular parts such as circulating blood, body fluids and clinical cell culture media from human samples [1]. The miRNAs can play an inhibiting function via basepairing complementary sequences of message-RNA (mRNA) to silence these genes by cleavage and repression mechanism, and therefore miRNAs are similar as small interfering RNAs (siRNA) by RNA interfering genes (RNAi). MiRNA come from regions of RNA transcripts with a forming short hairpin while siRNAs come from longer regions of double-strands. Because miRNAs only target mRNAs 6–8 nucleotides, combinatorial regulation is a special feature of miRNA, for example, a miRNA can target hundreds of different mRNA targets or a gene can be targeted by multiple miRNAs while a siRNA bind perfectly the genes of humans so that a siRNA can inhibit a gene completely [2].
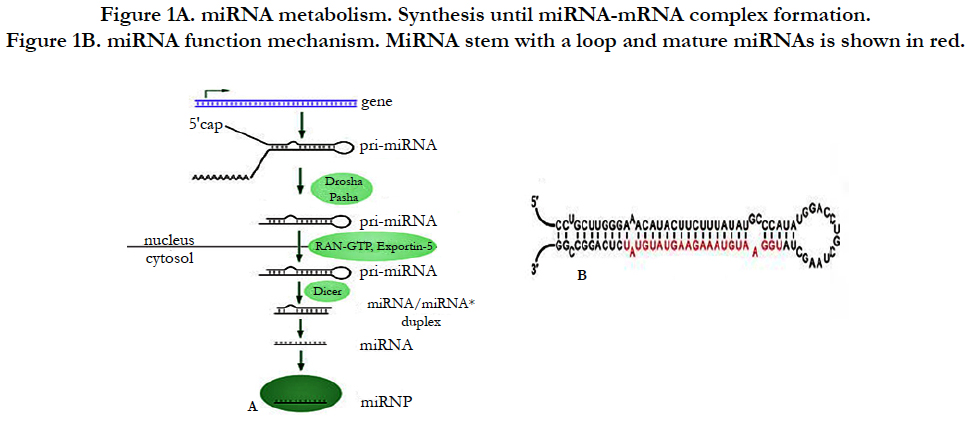
Following miRNA investigation, we have understood that miRNA is transcribed from sequences either from non-coding RNA or from coding genes [3]. Briefly as demonstrated in Figure-1A, after miRNA genes are transcribed by RNA polymerase II from the sequences, the transcripts will become hairpin loop, called as pri-miRNA. The resulting transcript is capped with a specially modified nucleotide at the 5’ end and a poly-A tail within 80 nucleotide RNA stem-loop. The double-stranded RNA (dsRNA) of the hairpins in a pri-miRNA is recognized by DGCR8 (nuclear protein, Digeorge Syndrome Critical region 8) which liberate the hairpin with stem called as a pre-miRNA (precursor-miRNA). Pre-miRNA hairpins are exported from the nucleus into cytoplasm by exportin-5 based on GTP energy support. Finally, the pre-miRNA hairpin (as shown Figure-1B) cleaved by the RNase III enzyme named as Dicer form a miRNAmiRNA duplex of 22 nucleotides. One strand of duplex results in miRNA-mRNA duplex to target mRNA by the formation of RNA-induced silencing complex (RISC) [4].
Figure 1B. miRNA function mechanism. MiRNA stem with a loop and mature miRNAs is shown in red.
In experimental study, miRNAs has been confirmed to target binding sites related to functions from tumor cells [5]. Chronic lymphocytic leukemia (CLL) is first disease to be discovered to relate with miRNA alteration [6]. The first miRNA mimic therapy entered the clinic for cancer therapy in 2013 [7]. After several years’ efforts, so many miRNAs have been uncovered to associate with tumor disease and tumorigenesis that now we call the miRNA profiles related with tumor diseases as “OncomiR, (or called as Oncomir)” [8]. Now, physicians and clinical scientists are going to study how to perform clinical sample to assay miRNA alterations, how to process miRNA detection for clinical miRNA analysis and diagnosis, how to use the miRNA profile to A: predict prognosis and monitor clinical response to treat tumor diseases; B: classify tumor diseases from miRNA biomarkers and C: apply for therapeutic purpose for tumor disease and tumorigenesis.
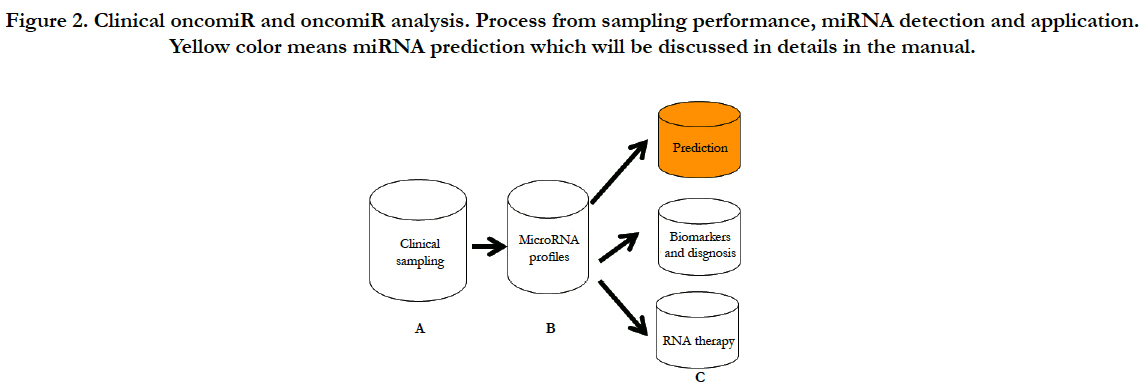
In order to clearly present clinical analysis and diagnosis from OncomiR files, here, I will conclude as shown at Figure-2 (A) clinical sampling for miRNA performance; (B) clinical miRNA detection with their analyses; (C) clinical application such as prediction of prognosis, diagnosis and therapy from the miRNA analysis; (D) in conclusion section, I will discuss challenges and future development of therapeutic targeting based on the clinical “oncomiR”.
Clinical Sampling
As discussed above, miRNA profile changes have been increasingly discovered to relate with tumorigenesis and tumor diseases. MiRNA profiles of specific genes or miRNA patterns of groups were found to be mainly associated with predictors of tumor prognosis and those of responses to chemotherapy. Some miRNAs also can be used for tumor classification according to miRNA biomarkers. Furthermore, some miRNAs are going to be used for clinical trial of miRNA treatment. In order to achieve accurate miRNA profile for the different application of clinical patients, here I first conclude different sampling methods for miRNA detection including non-tumor tissue sampling and tumor tissue sampling.
Non-tumor tissue processes for oncomiR analysis include body liquid specimens, cell free circulating miRNAs (cfmiRNAs), circulating tumor cell (CTC) and exosome. Non-tumor tissue processes are some very “popular” techniques for clinical scientists for downstream genomic performance, such as oncomiR performance.
In the early 2010, a report demonstrated oncomiR profiles from 12 body fluids, such as urine, sputum, mammary aspiration fluids, saliva and tears. Since then body fluidsused for miRNA detection have been increasingly reported [9]. Up to date, commercial products from several companies have largely reported to use them for miRNA performance[10].
Cell free circulating miRNAs (cfmiRNAs) are relative feasible biomarkers for tumor patients, which have been confirmed [11]. Now a lot of clinical evidences have demonstrated that cfmiRNAs are greatly related with tumor prognosis according to miRNA tumor biomarkers. For example, specific cfmiRNA profiles were discovered as prognosis predictor for patients’ survival and helicobacter pylori (HP) infection in esophageal adenocarcinoma. The research data classified 1075 miRNAs from 30 suffered patients and 30 healthy controls and finally uncover a combined cfmiRNA (low miR-3935 and high miR-4286) to relate with high-risk death (if it had >10- fold increased) [12]. Up to date, at least three company’s products (miRNeasy kit from Qiagen, miRVana PARIS kit from Ambion and total RNA isolation kit from Norgen Biotek) are often used for clinical cfmiRNA performance. The concentration of cfmiRNA is harvested 48.8 pg/μL from Qiagen kit, 29.3 pg/μL from Ambion kit and 11.7 pg/μL from Norgen kit. All the three products can be utilized for 200 ul serum with downstream PCR and miRNA-microarray although most of cfmiRNA products still require 2-4 ml blood serum and plasma for cfmiRNA process [13].
Exosomes are particles with 30-200 nanometer in diameter, which are isolated from almost all body liquids including serum, plasma, saliva, urine and so on [14]. Exosomes have shown that particles contain protein, lipid, DNA, mRNA and miRNA from tumor cells. Experimental studies have demonstrated that tumor cells exosomes involve in tumor cells growth, metastasis, decrease of immune response and enhancement of angiogenesis, because exosome can shed, a positive release, tens of thousands of vesicles from forms of multivesicular bodies (MVB) and direct budding at the plasma membrane, resulting in hundreds of billions of vesicles from tumor cells into plasma [15]. Clinically, at present, exosome mRNAs and miRNA (ex-miRNA) begin to be used to study global profiling of the mRNA/miRNA performance. Excitingly, a novel method of nanoscale fluorescence activated cell sorting (nanoFACS) has demonstrated a pure exosome isolation from fluids biopsy samples [16]. Now several companies are developing some products for exosome harvests, and thus ex-miRNA is easy to be applied for prediction, diagnosis and treatment of tumor diseases.
CTCs are cells shed into the circulation from a primary tumor location into distant organs from metastasis. Tumor cells of breast, prostate, lung, and colorectal cancer patients have been increasingly detected in circulation blood. We have been working in enriching clinical tumor cells from tumor patients and rare cells from genetic patients more than 20 years. Clinical evidence indicates that patients with metastatic lesions are more likely to have CTCs up to 1-10 CTCs in 1ml whole blood [17]. Isolating CTCs from around 10 ml blood are fundamental performance including gradient centrifuge, negative selection (CD45)/positive selection (CD326) called as enrichment-CTC [18].
Tumor tissue sampling includes clinical sampling in vitro; clinical sampling ex vivo and tissue level sampling with downstream analyses in silico for oncomiR analysis.
Clinical sampling in vitro for tumorcells isolation consists of laser-captured micro-dissection (LCM), magnetic cell separation (MACS) and flow-cytometric cell sorting (FACS) with downstream oncomiR analyses. LCM have very obvious advantages such as tumor cells morphology with their arrangement in tumor tissue on glass slides so that LCMs can enable us to select tumor-cells in vivo environment. Their combined Ab-based staining (IHC/ICC) and DNA/RNA based staining further increase the cell specificity from their biomarkers. MACS technique of clinical sampling in vitro is often used to sort cancer stem cells (CSC) and circulating tumor cells by cellsurface biomarkers such as CD133/CD34 for CSCs and EpCAM (CD326) for CTCs. MACS also support multi-step-labelling Abs, negatively and positively, to selectively harvest tumor cells and CTC which can increase its collection purity for a given tumor cells or CTC. FACS can isolate tumor cells or CSCs by a specific biomarker on the tumor cell surface which is as similar as MACS and intracellular biomarkers which is different from MACS technique. At present, multi-coloured FACS can one-step collect tumor cells and CTC by combined biomarkers from intracellular and extracellular parts [19].
Clinical sampling ex vivo for oncomiR analyses includes CSC or primary cancer cell culture with downstream miRNA analyses. Since 1994 we have reported 50 cases of primary tumor-cell culture for drug sensitivity assay, now we have routinely used the techniques to increase primary cell number with downstream clinical genomic analyses and drug screening [20].
In clinics, most specimens are directly frozen in liquid nitrogen from tumor removal. If the specimens are performed by oncomiR analysis at the tissue level, tumor tissues with downstream clinical oncomiR analyses in silico is a very important because the oncomiR data are mixed with different cells profiles from mixed cells at the tumor tissue [21].
After we understand clinical sampling process for miRNA performance, we need to determine which way is best to select patient samplings. If tumor samples are available such as surgically removal or biopsy, tumor cells sampling is first desirable. Because tumors are highly heterogeneous in tumor tissue, personalized medicine require specific and effective treatment. If physician need longitudinally monitor molecular change or clinicians need screen miRNA change for prediction of tumor and possible predictors for chemotherapy responses, liquid biopsy is a valuable choice because it is easily accessible and minimally invasive to determine effects [22].
Clinical MiRNA Detection with their Analyses
MiRNA detection methods for clinical specimens have been quickly developed by various technical arising. Due to miRNAs degrade more easily than mRNAs based on their length and RNase existence everywhere, previously, a great barrier of miRNA detection was an instability of miRNA. Now fresh specimens are immediately cooled with downstream performance under RNases-free condition and equipment, the miRNA degrade is obviously decreased by the new protocol. Here I present the methods as below which are often used in clinical laboratory.
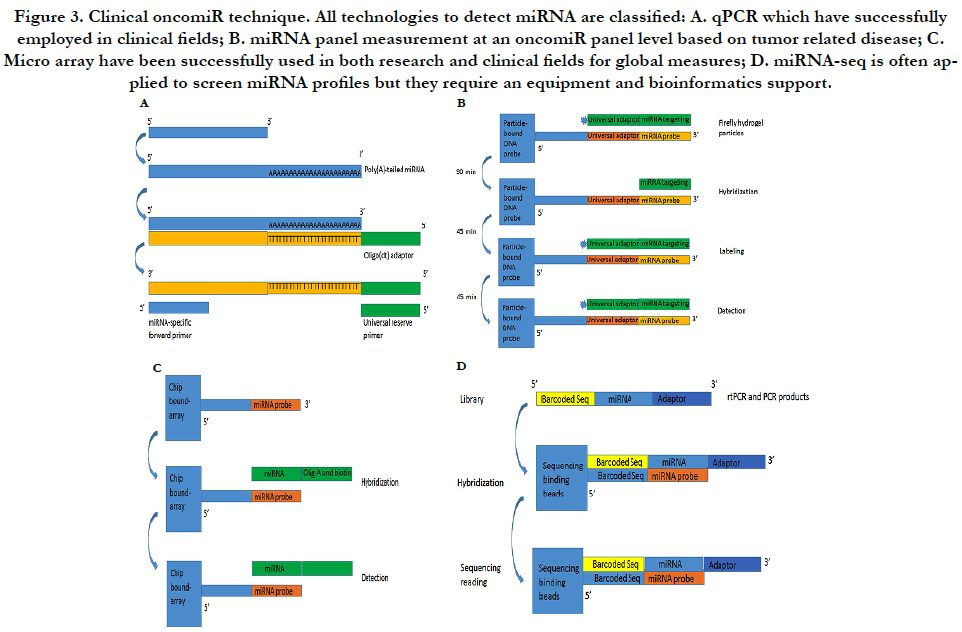
The miRNA qPCR assay has some special features including accurate quantification, higher sensitivity and relative inexpensive cost. They are often used for validating novel miRNAs if relatively small number of samples need to be assayed. Because the length of a miRNA is only 22 bp, two-step PCR processes consist of a modified rtPCR and a quantitative PCR as Figure-3A. The first step is to make the molecule longer by either incorporating a poly (A) tail or stem-loop structure while the second performance is initiated with an miRNA-specific primer and a stem-loop/poly (A) primer by SYBR® Green or a TaqMan® probe to detect the amplified product. In order to set up clinical performance, most of clinical laboratories use company products to process the both steps (modified and amplified) such as from Ambion or from Thermo-Scientific Inc or other companies [23].
Multiplex miRNA profiling is based on firefly particle technology to detect miRNAs. Detecting mechanism is to use some selected probes against target miRNAs in hydrogel particles. If some unknown miRNAs bind to these probes, the probes ligated to adaptor sequences can be detected as Figure-3B. An advantage is to run one set with multiple miRNAs and with a lot of samples without the labor intensive workflow such as miRNA extract. Moreover, data analysis is very simple so that most laboratories can perform the simple bioinformatics analysis. This technology can be used for both crude biofluids and miRNA samples. Currently, Abcam can support products to detect 10 μl of plasma/serum or 100 pg purified RNA by multiplex miRNA profile [24].
MiRNA arrays are to use thousands of probes with fluorescently labeled RNA samples on slides by photolithography to discover a potentially parallel change for all known miRNAs. We have used Exiqon with 1921 human miRNA to detect multiple miRNA targets (unpublished data) as Figure-3C. Although bioinformatics analysis can uncover a lot of putative miRNAs, the results still require experimental validation from these results of the miRNA arrays. Currently, several protocols and analysis tools are available for miRNA array performance. More important, because miRNA will involve in mRNA expression and because miRNA profile will be co-existed in mRNA microarray, the databases can be used to pair mRNA/miRNA-data that predict miRNA-mRNA targets based on their base sequence so that miRNAs of targets have been detected for integration mRNA/miRNA-expression information [25].
MiRNA-seq is high-throughput capability based on nextgeneration sequencing (NGS) platforms as Figure-3D. The most of clinical laboratories apply for TruSeq kits from Illumina Inc. by adding sequencing adapters to perform an RNA library and then isolate miRNAs by size of small RNA products. MiRNAseq is an ideal method but it cannot be quantified miRNA levels as qPCR and multiplex miRNA panel. Furthermore, miRNA-seq can discover new miRNAs and their isoforms although it need a bioinformatics platform to support [26].
After we comprehend clinical miRNA detection for oncomiR process, clinical scientists require to choose miRNA detection methods including downstream analysis. The selection is relied on clinical purpose and miRNA sample size. For example, if miRNA assayed as one or few with very few miRNAs samples require further confirmation, qPCR assay is first option while Firefly particle technology is more suitable for multiplex miRNA panel with relatively large number of samples. MiRNA-seq or miRNAarray will be very good candidates for scientists to screen new miRNA alteration. Finally, miRNA profiles performance also should be considered their downstream analysis such as skilled bioinformatics scientists and available tools.
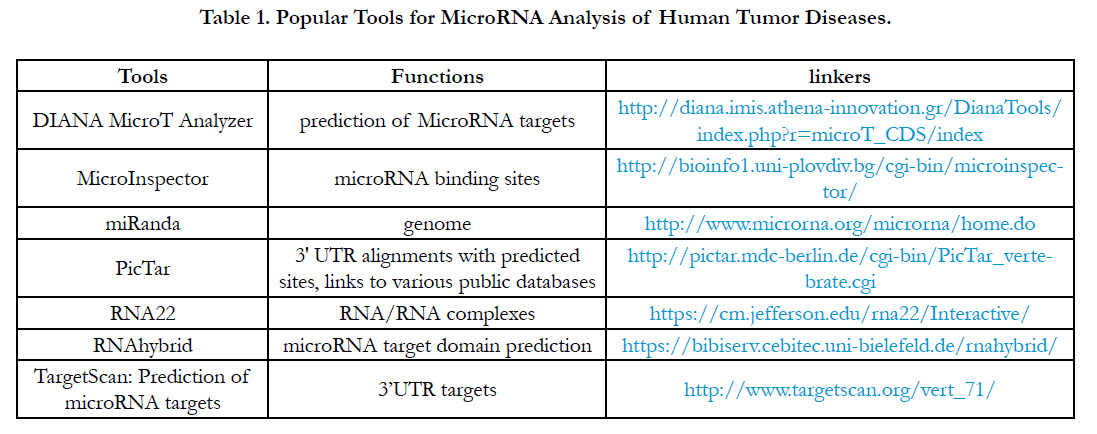
After we harvested a miRNA profile from miRNA array or miRNA-seq or multiplex miRNA profiles, theoretically, these miRNA profiles require a powerful high-throughput tool to study a regulation by miRNA-mRNA regulation or miRNA-pathway from the miRNA profiles [27]. Clinically, understanding the tools related tumor disease can be applied to the analysis of miRNA data resulting in a further application for tumor diseases. Although so many software tools have been developed, here I just introduce the software tools which can be often used for miRNA analysis for tumor diseases as Table 1.
Clinical Application
In early periods, scientists mainly focused on study of miRNAbased cancer biology such as controlling expression of their target mRNAs for tumor growth, invasion and angiogenesis in clinical fields. After several years’ efforts, miRNA profiles have been increasingly utilized for clinical patients such as predict tumor prognosis and monitor response of chemotherapy and RNA therapy.
After tremendous studies of the miRNAs alterations related to tumor disease, oncomiR profiles has given physicians a great support to predict prognosis/progress of tumor disease and drug responses for different therapies. Now, although gene expression level of mRNA and protein still play a key role in personalized medicine, personalized medicine requires more personal information to support the new fields including miRNA change, SNP-based GWAS and methylation-based epigenetics alterations. For example, miRNAs may contribute much more message forprediction of prognostic indicators than those of mRNAs. The foremost, miRNA profiles will give clinicians more reliable information because miRNA degradation have been resolved.
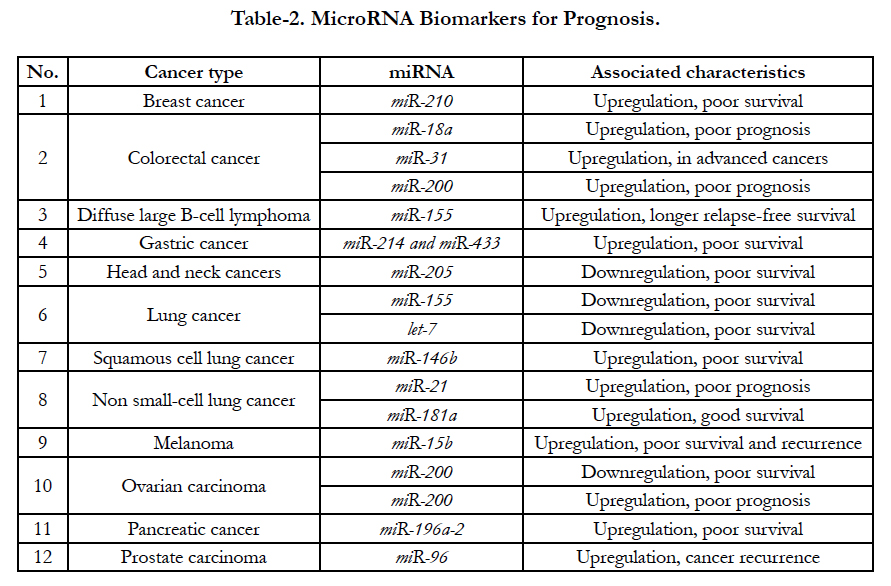
According to recent clinical assays, several miRNAs increase and decrease have been demonstrated in some special tumors [28]. For examples, up-expressed miR- 210 are related with poor survival from breast cancer patients; up-expressed miR-18a, miR-200 and miR-31 are also associated with poor prognosis from colorectal cancer while downregulated miR-155 and let-7 are related with poor survival from lung tumor [29]. All relationship between up- and down-regulation are shown at Table-2. Recently, clinical assays demonstrated that miR- 21 enhanced (oncogenic miRNA) and let-7 (onco-suppressor miRNA) decreased in tumor diseases are the most miRNAs biomarkers for miRNAs alterations. The oncogenic miR-21 is overexpressed in many tumor diseases, including hepatocellular carcinoma, lung cancer, stomach cancer, colorectal cancer, breast cancer, prostate cancer, and glioblastoma as well [30].
MiRNA may be is a good predictor to monitor response of chemotherapy although all profiles are still in the study. Recent studies have confirmed that some miRNAs can guide therapy decisions. For instances, after a successful investigation for miR-34 modulators in Wnt and TGF pathway, miR-34a inducing-apoptosis and inhibiting-cell proliferation have been discovered by clinical data. Higher miR-34 regulation is effective indicator for clinically effective therapeutics [31]. Moreover, let-7 family of miRNAs also are important regulatory molecules in lung cancer, esophageal cancer and colon cancer. KRAS is an important oncogene in the etiology in lung adenocarcinoma, esophageal cancer and colon cancer, which was discovered by downstream target of let-7. Clinically, the data have supported that let-7-LCS6 polymorphism can predict the efficacy of anti-EGFR-based therapy in patients with metastatic colorectal cancer [32]. Another example of altered target sites is miR-191 in the 30UTR of MDM4 by the presence of SNP34091, which show to affect chemo-sensitivity in ovarian cancer. In chronic myeloid leukemia (CML) with the BCR-ABL rearrangement, miR-451 can interfere imatinib treatment (selective BCR-ABL inhibitor) after miR-451 levels were confirmed to inversely correlate with BCRABL levels [33]. All current miRNA predictor of responses to chemotherapy and molecular therapy are listed in Table 3.
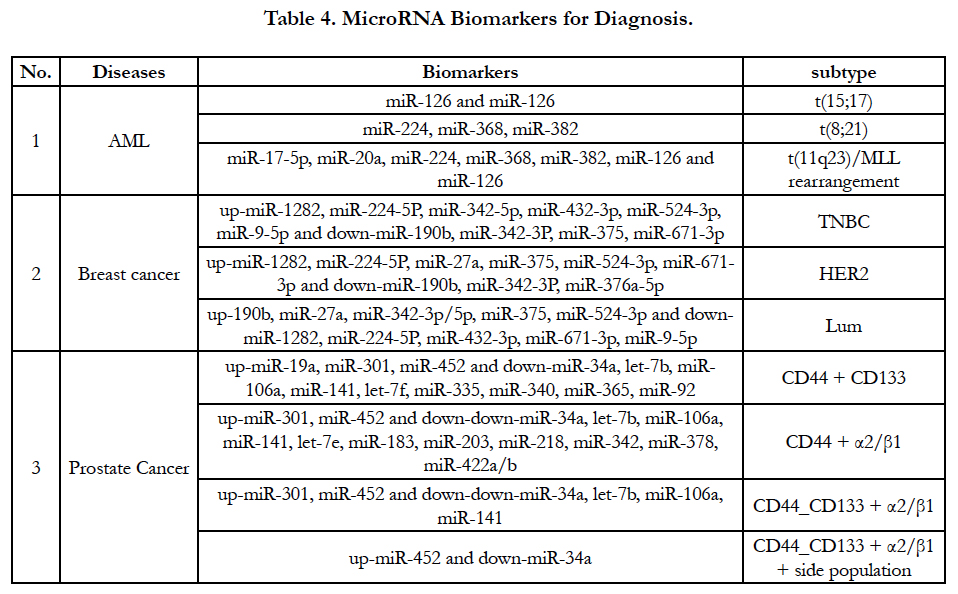
MiRNA expression level have been increasingly used for a signature of the tumorigenesis, tumor classification and subtype. An accurate result of miRNA expression level supports tumor classification and subtype based on gene translocation/rearrangement and cell origin. For example, acute myeloid leukemia (AML) have at least three types of translocation and MLL rearrangement [t(15;17), t(8;21) and t(11q23)/MLL rearrangement]. Accordingly, three parallel miRNA alterations will be demonstrated in these kinds of genes’ translocation and rearrangement as Table-4 [34]. In breast cancer there are three types of cell origins such as epithelial origin (luminal), basal-like (myoepithelial origin) and human epidermal growth factor receptor 2 (HER2). MiRNAs can classify breast cancer according to tumorigenesis and receptors of breast cancer [35]. MiRNAs profiles also define prostate cancer subtype based on cell origin such as CD44 source, CD133 source, α2/ β1 protein and side population [36]. Because miRNA largely exist in body liquid such as sera, plasma and other bod liquid, the detection of miRNAs may be can partially substitute mRNA/ protein detection for diagnosis, prognosis assessment and cancer progression prediction. Excitingly, after cfmiRNA and exome are screened in different cancer types shown in Table 2, Table 3 and Table 4, detection of the miRNAs from patients’ blood should be greatly developed because non-invasive method will be easy to be accepted by patients and physicians.
As discussed above, every miRNA has multiple target sites in different genes while mRNAs have few conserved bindingsequences that are predicted to interact with miRNAs. Based on the miRNA mechanism, miRNA-based therapeutics have a very obvious advantage such as miRNAs as therapeutic agents to target multiple genes in networks. Of course, one miRNA targeting multiple genes also can cause more toxic side-effects due to multiple targets in a network system. A challenge is how to use, effectively and specifically, delivery system for miRNA therapeutics purpose. Effective miRNA delivery system is relied on miRNA stability in circulation system because unmodified RNAs will be quickly cleared from circulating blood within 30 minutes. Now chemically modified oligonucleotides have been largely used for the miRNA stability such as locked nucleic acid (LNA) oligonucleotides, phosphorothioate containing oligonucleotides, 2’–O-methyl-(2’–O-Me) or 2’–O-methoxyethyl-oligonucleotides (2’–O-MOE), peptide nucleic acids (PNA), fluorine derivatives (FANA and 2’-F). MiRNA specificity deliver is based on accurate delivery of miRNAs [37]. Currently, most delivery of miRNAs are not specific to tissue such as cholesterol moiety, liposomenanoparticles and polycationic liposome-hyaluronic acid (LPH) nanoparticles. Other non-specific delivery systems are polyethylenimine (PEI)-based systems, dendrimers, poly (lactideco- glycolide) (PLGA) particles, protamine, atelocollagen and silica-based nanoparticles [38]. Although non-specific delivery system is going to study, some specific tissues delivery systems are going to be developed by either binding tumor-specific ligands to RNA-containing-particle by using HER-2 and CD44 or tissuespecific expression for some specific tissue or cells [39].
Theoretically, miRNA treatment consist of miRNA inhibition therapy for oncogenic miRNA (called as oncogenic miRNA) when it is highly expression and miRNA replacement therapy (named as mimic-miRNA) for tumor suppressor miRNA in which the miRNA is repressed. MiR-122 has been demonstrated for the replication of hepatitis C virus (HCV) by binding of miR- 122 to the 5′ noncoding region of the HCV genome in liver cells. Anti-miR-122 is the first miRNA-based treatment for hepatitis C virus in patients. Now the candidates of miRNAs include miR-21 and miR-155 (oncogene) and miRNA-29 family (miR-29a, miR- 29b, and miR-29c), miR-29 family and miR-10b (antagomiRs). On the other hands, tumor suppressor miRNAs for mimic replacement therapy such as let-7 and miR-34 have increasingly reported in animal model. Encouragingly, a liposomal miR-34 mimic replacement therapy, called as MRX34, is investigated for human tumor disease by Mirna Therapeutics Inc [40].
Conclusion
MiRNA assay is a new clinical tool to detect miRNA alterations, which can be used for monitor prognosis of tumor disease, predict responses for different therapies and future RNA therapy. Although detecting oncomiR profiles has been quickly developed into clinics during last several years, several questions still should be addressed. Here the four issues listed below should be resolved in the next periods:
In clinical genomic diagnosis, we have discovered a limitation for genomic application if we have not set up a successful diagnosis model. If we performed specificity and sensitivity from the miRNA profile, effects of diagnosis and treatment will be improved from oncomiR profiles.
Recent development of cancer research has enabled scientists to use different oncomiR profiles for clinical application. If we use the data for clinical diagnosis and treatment of patients, we need to further confirm the oncomiR data.
Genomics-based diagnostic tests have been greatly developed in direct therapeutic interventions. In order to improve clinical practice from clinical oncomiR analysis, quantitative network supporting clinical oncomiR analysis have been developed to provide personalized medicine for patients. As I discussed above, in order to improve effects for miRNA diagnosis, we are going to develop quantitative network from oncomiR data to define specific and sensitive therapeutic targeting for the aim of patient’s miRNA diagnosis and treatment.
After oncomiR profiles are discovered, miRNAs profiles, currently, have been only used for tumor prediction, prognosis and diagnosis for tumor subtypes. Because genomic regulation is systemic mechanism, we need combine other genomics, for example, miRNA-mRNA combination related with tumors should be extended into drug discovery for personalized therapies; miRNA-SNP combination profiles related with responses of chemotherapy should be developed into personalized medicine. In the near future, physician should combine the data at different level (DNA, RNA, protein level with connection with Ab, drugs and other molecules) for the precision medicine.
Acknowledgments
Under the support of Dr. H. D. Preisler, we had set up the method to analyze clinical genomic analysis including single-cell genomic profiles of tumor cell from solid tumors and leukemia. Now we are going to use new genomic platform to continue working in epigenetic aberrance. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation.
References
- Mraz M, Malinova K, Mayer J, Pospisilova S (2009) MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 390(1): 1-4.
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell. 115 (2): 209–16.
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 10(12): 1957–66.
- Lin SL, Chang D, Ying SY (2005) Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 356: 32–8.
- Spiegl-Kreinecker S, Pirker C, Filipits M, Lotsch D, Berger W, et al., (2010) O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro-oncolo. 12(1): 28–36.
- Mraz M, Pospisilova S (2012) MicroRNAs in chronic lymphocytic leukemia: from causality to associations and back. Expert Review of Hematology. 5(6): 579–81.
- Farooqi AA, Fayyaz S, Shatynska-Mytsyk I, Javed Z, Jabeen S, et al., (2016) Is miR-34a a Well-equipped Swordsman to Conquer Temple of Molecular Oncology? 87(3): 321-34.
- Mohammadi A, Mansoori B, Baradaran B (2017) Regulation of miRNAs by herbal medicine: An emerging field in cancer therapies. Biomed Pharmacother. 86: 262-270.
- Schmidt B, Rehbein G, Fleischhacker M (2016) Liquid Profiling in Lung Cancer - Quantification of Extracellular miRNAs in Bronchial Lavage. Adv Exp Med Biol. 924: 33-37.
- Wang Y, Liang Z, Gao Y, Zhai D, Rao Q, et al., (2015) Factors influencing circulating MicroRNA level in the studies of hepatocellular carcinoma biomarker. Neoplasma. 62(5): 798-804.
- Fumagalli C, Bianchi F, Raviele PR, Vacirca D, Fusco N, et al., (2017) Circulating and tissue biomarkers in early-stage non-small cell lung cancer. Ecancermedicalscience. 11: 717.
- Zhai R, Wei Y, Su L, Wain John C, Kulke MH, et al., (2015) Whole-miRNome profiling identifies prognostic serum miRNAs in esophageal adenocarcinoma: the influence of Helicobacter pylori infection status. Carcinogenesis. 36(1): 87-93.
- Roy S, Soh JH, Ying JY (2016) A microarray platform for detecting diseasespecific circulating miRNA in human serum. Biosens Bioelectron. 75: 238-46.
- Shao Y, Shen Y, Chen T, Xu F, Chen X, et al., (2016) The functions and clinical applications of tumor-derived exosomes. Oncotarget. 7(37): 60736-60751.
- Janas AM, Sapoń K, Janas T, Stowell MH, Janas T (2016) Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochim Biophys Acta. 1858(6): 1139-51.
- Nolan JP (2015) Flow Cytometry of Extracellular Vesicles: Potential, Pitfalls, and Prospects. Curr Protoc Cytom. 73: 13.14.1-16.
- Mego M, Gao H, Cohen EN, Anfossi S, Sanda T, et al., (2016) Circulating Tumor Cells (CTC) Are Associated with Defects in Adaptive Immunity in Patients with Inflammatory Breast Cancer. J Cancer. 7(9): 1095-104.
- Singer BD, Mock JR, D'Alessio FR, Mandke P, Agarwal NR, et al., (2016) Flow-cytometric method for simultaneous analysis of mouse lung epithelial, endothelial, and hematopoietic lineage cells. Am J Physiol Lung Cell Mol Physiol. 310(9): L796-801.
- Kitayama J, Emoto S, Yamaguchi H, Ischigami H, Seto Y, et al., (2015) Flow Cytometric Quantification of Intraperitoneal Free Tumor Cells is a Useful Biomarker in Gastric Cancer Patients with Peritoneal Metastasis. Ann Surg Oncol. 22(7): 2336-42.
- Li B, Tong SQ, Zhang XH, Lu J, Gu JL, et al., (1994) A New Experimental and Clinical Approach of Combining Usage of highly Active TIL and Highly Sensitive Antitumor Drugs for the Advanced Malignant Tumor. Clin Med J. 107(11): 803-807.
- Yang IS, Son H, Kim S, Kim S (2016) ISOexpresso: a web-based platform for isoform-level expression analysis in human cancer. BMC Genomics. 17(1): 631.
- Zhang XX, Deng LH, Chen WW, Shi N, Jin T, et al., (2017) Circulating microRNA 216 as a Marker for the Early Identification of Severe Acute Pancreatitis. Am J Med Sci. 353(2): 178-186.
- Choi JL, Kao PF, Itriago E, Zhang Y, Hoss AG, et al., (2017) miR-149 and miR-29c as candidates for bipolar disorder biomarkers. Am J Med Genet B Neuropsychiatr Genet. doi: 10.1002/ajmg.b.32518.
- Urquidi V, Netherton M, Gomes-Giacoia E, Serie DJ, Rosser CJ, et al., (2016) A microRNA biomarker panel for the non-invasive detection of bladder cancer. Oncotarget. 7(52): 86290-86299.
- Esser JS, Saretzki E, Pankratz F, Engert B, Moser M, et al., (2017) Bone morphogenetic protein 4 regulates microRNAs miR-494 and miR-126-5p in control of endothelial cell function in angiogenesis. Thromb Haemost. 10.1160/TH16-08-0643.
- Siska C, Kechris K (2017) Differential correlation for sequencing data. BMC Res Notes. 10(1): 54.
- Chandrasekaran S, Bonchev D (2016) Network analysis of human postmortem microarrays reveals novel genes, microRNAs, and mechanistic scenarios of potential importance in fighting huntington's disease. Comput Struct Biotechnol J. 14: 117-130.
- Raychaudhuri M, Bronger H, Buchner T, Avril S, Weichert W, et al., (2017) MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. doi: 10.1007/s10549-017-4132-9.
- Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, et al., (2016) MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 69(14): 5776-83.
- Feng YH, Tsao CJ (2016) Emerging role of microRNA-21 in cancer. Biomed Rep. 5(4): 395-402.
- Zhang DG, Zheng JN, Pei DS (2014) P53/microRNA-34-induced metabolic regulation: new opportunities in anticancer therapy. Mol Cancer. 13: 115.
- Saridaki Z, Weidhaas JB, Lenz HJ, Teipar S, Zhang W, et al., (2014) A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage etuximab/panitumumab monotherapy. Clin Cancer Res. 20(17): 4499-510.
- Scholl V, Hassan R, Zalcberg IR (2012) miRNA-451: A putative predictor marker of Imatinib therapy response in chronic myeloid leukemia Leuk Res. 36(1): 119-21.
- Lin X, Wang Z, Wang Y, Feng W (2015) Serum MicroRNA-370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int J Clin Exp Pathol. 8(11): 14658-66.
- Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Horiquchi J, et al., (2017) Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet. 62(1): 15-24.
- Liu C, Kelnar K, Vlassov AV, Brown D, Dean G, et al., (2012) Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 72(13): 3393-404.
- Gandellini P, Profumo V, Folini M, Zaffaroni N (2011) MicroRNAs as new therapeutic targets and tools in cancer. Expert Opin Ther Targets. 15(3): 265-79.
- Jain CK, Gupta A, Dogra N, Kumar VS, Sharma SK, et al., (2014) Micro-RNA therapeutics: the emerging anticancer strategies. 9(3): 286-96.
- Kosovrasti VY, Nechev LV, Amiji MM (2016) Peritoneal Macrophage-Specific TNF-α Gene Silencing in LPS-Induced Acute Inflammation Model Using CD44 Targeting Hyaluronic Acid Nanoparticles. Mol Pharm. 13(10):3404-3416.
- Beg MS, Brenner AJ, Sachdev J, Borad M, Smith S, et al., (2016) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs.