Expression of Tropomyosin 2 Gene Isoforms in Human Cardiac Tissue
Syamalima Dube1, Santhi Yalamanchili1, Joseph Lachant1, Lynn Abbott1, Patricia Benz1, Dipak K Dube1, Bernard J Poiesz1*
1 Department of Medicine, Upstate Medical University, 750 East Adams Street, Syracuse, NY, USA.
*Corresponding Author
Bernard J Poiesz,
Department of Medicine,
Upstate Medical University,
750 East Adams Street,
Syracuse, NY, USA.
Tel: 315-464-8231; Fax: 315-464-8255
E-mail: poieszb@upstate.edu
Article Type: Research Article
Received : June 04, 2014; Accepted: June 23, 2014; Published: June 25, 2014.
Citation: Poiesz BJ et al., (2014) Expression of Tropomyosin 2 Gene Isoforms in Human Cardiac Tissue. Int J Cardiol Res. 1(3), 9-14. doi: dx.doi.org/10.19070/2470-4563-140003
Copyright: Poiesz BJ© 2014. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Previous studies have shown that although the transcript levels of TPM1α and TPM1κ are expressed in human hearts in comparable levels, the level of TPM1α protein is ~90%. The proteins of TPM1κ and TPM2α are about 5% of the total sarcomeric TM. The TPM2 gene is known to generate three alternatively spliced isoforms, which are designated as TPM2α TPM2β TPM2γ. The expression level of TPM2β and TPM2γ in human hearts is unknown. Using a series of primers pairs and probes for RNA PCR, we found that both TPM2α and β but not γ were expressed in fetal and adult heart tissue, with about the same amounts of each isoform in fetal hearts and more β than α in adult hearts. Four new isoforms of TPM2 RNA were identified (TPM2δ-η). Most of these were present in very small amounts in both the fetal and adult hearts with the exception of TPM2ξ, which was present at about 40% of the level of TPM2α in adult heart tissue. Western blot analyses using a series of anti-tropomyosin antibodies indicate that TPM2 protein is present in both fetal and adult hearts at about the same levels as TPM1κ and much less than TPM1α. We are unsure about the expression of TPM2δ, TPM2ζ, and TPM2η proteins in fetal and adult human hearts. The exact function of these new TPM2 isoforms in heart and their role(s) in cardiac disease remain to be elucidated.
2.Introduction
3.Materials and Methods
4.Results
4.1.Cloning and sequencing of four novel TPM2 isoforms
4.2.Expression of transcripts of the novel TPM2 isoforms
4.3.Western blot analyses of the extracts from fetal and adult human hearts with various anti-tropomyosin antibodies
5.Discussion
6.Acknowledgement
7.References
Key Words
Alternatively Spliced; RT-PCR; Cloning & Sequencing; Westernblot Analyses.
Introduction
Tropomyosin (TM) is a component of myofibrils, the contractile apparati of striated muscle cells [1-6]. Recent associations of TM mutations with various myopathies in humans have sparked renewed interest in the structural/functional relationships in TM [7- 10]. Vertebrate TMs are expressed from four tropomyosin genes designated as TPM1, TPM2, TPM3, and TPM4 [1-6] except for fish where there are six TPM genes [11]. More than twenty distinct isoforms are generated via a complex pattern of alternative RNA splicing and alternative promoters. However, the functional significance of this isoform diversity is poorly understood. Also, it is not clear whether specific isoforms are required for assembly and integration into distinct actin-containing structures ( Helfman et al. (1999). Helfman et al. using GFP tagged TM have shown that both muscle and nonmuscle isoforms can be incorporated into the I-band of neonatal rat cardiac myocytes (NRCs). However, the functional role of each of the non-muscle isoforms in cardiac contractility is yet to be elucidated.
TPM1 gene in vertebrates was known to form nine different alternatively spliced TM isoforms. We were the first to report the tenth TPM1 isoform, which happens to be sarcomeric isoform and is designated as TPM1κ [13,14,15]. Cooley and Bergstrom (2001) also reported several new isoforms of the TPM1 gene [16]. The TPM2 gene is known to produce three TM isoforms, TPM2α (muscle isoform), TPM2β (TM-1 or nonmuscle isoform), and TPM2γ (in HeLa cells) via alternate splicing. To the best of our knowledge, an extensive analyses of isoform diversity of the TPM2 gene has not yet been reported. In this study, we have decided to explore this issue and search for the presence of novel TPM2 RNA and protein expression in adult and fetal human hearts. We have detected, cloned, and sequenced four new alternatively spliced isoforms of the TPM2 gene from human adult and fetal hearts hearts. These are designated as TPM2δ, TPM2ε, TPM2ζ, and TPM2η. While preparing this manuscript, predicted sequences of several potential TPM2 isoforms including our TPM2ε and TPM2η have been submitted in the Genbank in 2014. Hence, our claim that this is the first report of the expression of TPM2δ, TPM2ε, TPM2ζ, and TPM2η in human cardiac muscles is well justified. Furthermore, using Western blot analyses and a variety of anti-tropomyosin antibodies, we have made an effort to detect the protein expression of all TM isoforms in fetal and adult human hearts.
Materials and Methods
Human adult and fetal cardiac total RNA was obtained from Zyagen (San Diego, CA) and similar protein samples were obtained from Imgenex (San Diego, CA). The samples represent in each case material from one adult heart and 5 fetal hearts.
Human skeletal muscle protein was obtained from Imgenex. The non-malignant human breast epithelial cell line MCF10A and the normal human B-lymphocytic cell line HCC 1143 (BL) were obtained from ATCC (Manassas, VA). Total cellular RNA and protein were prepared from the cell lines, as previously described. For RT-PCR, 0.5 µg of RNA in a total volume of 40 µl was used to synthesize cDNA with SuperScriptR II (Life Technologies, Grand Island, NY) and oligo-dT primers following the manufacturer’s specifications. For each PCR 3 µl of cDNA was used, GAPDH housekeeping gene RNA was amplified as previously described. RT-PCR for TPM2 RNA isoforms utilized the following primers located in different exons of the complete TPM2 RNA transcript (Fig 1).
exon 1: 5´ - ATGGACGCCATCAAGAAGAA - 3´ (+)
exon 9a: 5´- CTTGTACTTCATCTTCTGGGCATAG - 3´ (-)
exon 9b: 5´ - CACATGCAGTGGTGAATCA - 3´ (-)
exon 9d: 5´ - TGGGGCTGGCCCTCACAGGTT - 3´ (-)
TPM1α and TPM1κ primers and probes were as previously described.
Amplified DNA was detected using the following probes.
exon 5: 5´ - AGAGGGCTGAGGTGGCCGAGAGCCG - 3´ (+)
exon : 5´ - CCTGGCACTGAGCCCACCCACAA - 3´ (+)
cDNA was amplified and detected using Southern blot hybridization,as previously described.
Amplified products were ligated and cloned into the TA cloning vector (Life Technologies) following our published protocol (13). Positive clones were identified using the above specific probes,as previously described. Vectors were grown in E. coli, and the DNA was extracted using Qiagen mini-prep kit (Valencia, CA). The isolated DNA was then sequenced (Cornell University Life Science Core Laboratories Center, Ithaca, NY). Each colony was sequenced twice in both directions. The relative amounts of eachTPM2 isoform was determined by comparing their Southern blot hybridization signals to that of the GAPDH of reference housekeeping gene signal, as previously described. The frequency of various TPM2 isoforms was determined by sequencing DNA from 100 different colonies from fetal heart cDNA, and 5 different colonies from adult heart cDNA originally amplified with the TPM2 exon 1 and exon 9d primers.
LDS sample buffer and sample reducing agent (Life Technologies) were added to the protein extracts, and SDS PAGE was carried out following our published protocol. About 10 μg of protein extracts from human adult and fetal hearts, human skeletal muscle and the cell lines MCF 10A and HCC 1143 (BL) were heated at 70°C and subsequently loaded into Novex NUPAGE 4-12% BIS-TRIS gels in MOPS running buffer with antioxidant added (Life Technologies), and run for 1 h at 200 V in a Cell Sure Lock Mini-Cell apparatus (Life Technologies). The gels were transferred to nitrocellulose membranes using the same apparatus, under transfer buffer (Life Technologies) supplemented with methanol and antioxidant. Ponceau reversible staining was done to test loading consistency and transfer efficiency. The blots were blocked in 5% dry milk powder (Nestle HealthCare Nutrition, Inc., Florham Park, NJ), in TBST (10x TBS, 0.05% Tween-20) overnight at 48°C. Primary antibody incubations were carried out for 1 h at room temperature. Blots were washed in TBST after both primary and secondary antibody incubations. Chemiluminescence was accomplished using ECL detection reagents (Amersham, Piscataway, NJ) and exposing the blots to x-ray film (Fuji Film, FP-3000B), following the manufacturer’s protocol. Antibodies were diluted in the 5% milk powder blocking solution. Primary antibodies included TM311 (Sigma-Aldrich, St. Louis,MO) anti-TPM1κ, CH1 (Sigma) and the anti-TPM2 exon 6a, 187-206 (a kind gift of G.L. Prasad, Department of General Surgery and Cancer Biology, Wake Forest University School of Medicine, Winston-Salem, NC). Secondary antibodies were goat anti-rabbit immunoglobulin HRP and sheep anti-mouse immunoglobulin HRP (GE Healthcare).
Results
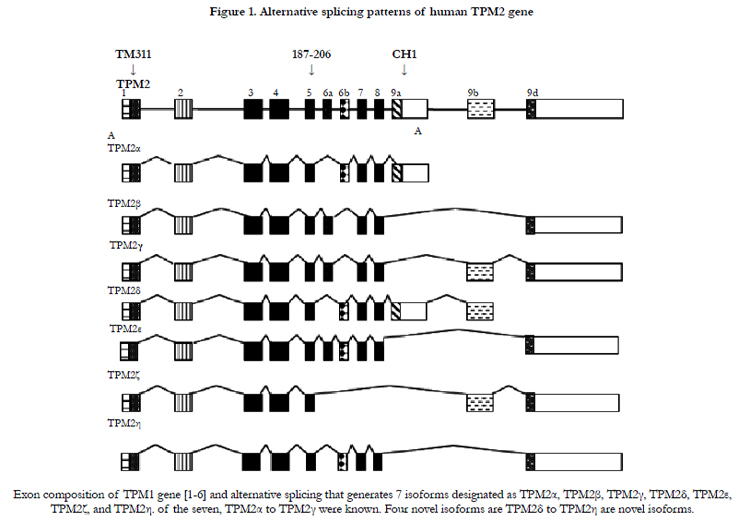
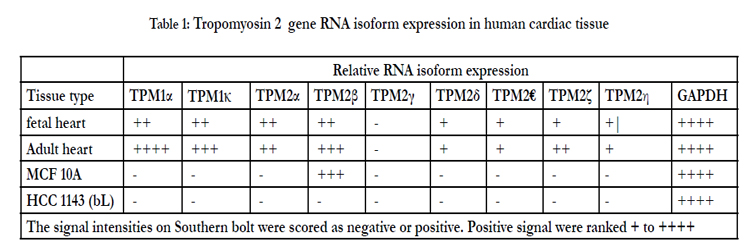
Both human fetal and adult heart tissue expressed TPM2α and TPM2β but not TPM2γ (Table 1). Four novel TPM2 RNA isoforms were detected by RT-PCR, which we have labeled TPM2δ, TPM2ε, TPM2ζ, and TPM2η (Table 1). These were present in both the adult and fetal heart tissues. The organization and exon content of each of the above TPM2 isoforms is shown in Figure 1. The signal intensities of the Southern blots of each isoforms relative to GAPDH are shown in Table 1. The nucleotide as well as the deduced amino acid sequences of these isoforms are presented in Figure 2. TPM2δ encodes 284 amino acid residues, which are identical with TPM2α. The only difference between TPM2α and TPM2δ is the 3’-UTR. The transcript of the TPM2α ends at exon 9a whereas the transcript of TPM2δ includes exon 9b due to the novel splicing. The splice variant TPM2ε contains both exon 6a and 6b. The transcript stops at exon 9b. This isoform encodes 213 amino acid residues due to the formation of a premature stop codon after exon 6a and at the beginning of exon 6b. The nucleotide as well deduced amino sequences of TPM2ε are identical with those of the predicted splice variant X7 (XM_005251571). TPM2ζ isoform encoding 240 amino acid residues contains exon 1, 2, 3, 4, 5, 9b, and 9d and lacks both 6a and 6b (Figure 2C). The other novel isoform TPM2η encodes 284 amino acid residues, which are identical with TPM2β except for exon 6. TPM2β contains exon 6a whereas, TPM2η contains exon 6b. The amino acid sequence of this isoform is identical with the predicted TPM2 variant X6 (XM_005251570).
TPM2α and β were equally expressed in fetal heart, while TPM2β was expressed a bit more in adult hearts. TPM2δ, TPM2ε, and TPM2η gave weak signals for adult heart tissue; they and TPM2ζ gave weak signals in the fetal heart tissue. However, TPM2ζ gave a stronger signal in adult heart. In order to be more precise with the relative frequencies of the TPM2 isoforms β through η, we amplified with the common exon 1 and exon 9d primers and analyzed 100 and 20 colonies of cloned cDNA from the fetal heart and adult heart tissue, respectively (Table 2). It should be remembered that these primers would not amplify TPM2α (Figure 1). As can be seen, 96% of the colonies in the fetal heart were TPM2β, none were TPM2γ and the other isoforms were present in 1 colony each. In the adult heart 85% of the colonies were TPM2β and 15% were TPM2ζ. As can be seen, in the adult heart tissue, there is much more TPM1α than TPM1κ, TPM2α or TPM2β RNA, but TPM2α and TPM2β RNA is present at about the same or slightly greater amounts as TPM1κ (Table 1). In the fetal heart tissue TPM1α is, again, the more prevalent isoforms while there is slightly more TPM1κ than TPM2α and TPM2β.
Table 3 shows Western blot data using the antibodies TM311, anti-TPM1κ, CH1 and TPM2 187-206. The various bands observed and, the known or putative TPM1, TPM2, and/or TPM3 isoforms that should be observed with those antibodies at a particular molecular weight are indicated. TPM1α was the dominant tropomyosin protein in both adult and fetal hearts with much lower levels of TPM1κ present in both as well. TPM2 protein was present in both, as well, in amounts roughly equivalent to TPM1κ. No TPM2γ or TPM2ε or TPM2ζ protein expression was observed. Given the low level TPM2η RNA observed then, by deduction, there is definite expression of TPM2β protein in both fetal and adult hearts at the level of TPM1κ. Given that we have no specific TPM2κ antibody and that TPM2α protein comigrates with TPM1κ, we cannot definitively state that TPM2α is expressed.
Discussion
We have identified and characterized four different TPM2 splice variants in human fetal and adult hearts. Due to a lack of isoform- specific antitropomyosin antibodies, we are unsure about the protein expression from these splice variants in human hearts. The muscle type novel isoform TPM2δ and the known TPM2α isoform have distinctly different 3’-UTRs but encode the identical protein containing 284 amino acid residues. Currently, we do not know whether these two isoforms are expressed in the same cardiac tissues for example, myocytes, or fibroblasts or any other cell types. As the 3’-UTRs are known to play a critical role in the translation of a variety of mRNAs [17-20], we can speculate that these two mRNAs may be differentially translated in different cardiac cell types for example myocytes, fibroblasts, etc. depending on their specific requirements. Before drawing a definitive conclusion, it is an essential pre-requisite to determine the exact expression patterns of these two mRNAs in different cardiac cell types.
In a classical sense, three of the four novel isoform TPM2e,TPM2z, and TPM2η are non-muscle isoforms because none of them contain exon9a that encodes the troponin binding domain of sarcomeric TMs. Troponins are an indispensable components of thin filaments in sarcomere in muscle tissues [1-5]. However, the permanent cellular constituents of the heart include myocytes, cardiac fibroblasts, endothelial cells, and vascular smooth muscle cells [2]. Therefore, these novel TPM2 isoforms may be expressed in cardiac cell types other than cardiomyoctes. Current dogma states that fibroblasts make up the largest cell population of the heart [21]. Again, cardiac fibroblasts play a critical role in maintaining normal cardiac function, as well as in cardiac remodeling during pathological conditions such as myocardial infarct and hypertension. Hence, it is not illogical to speculate that some of these isoforms may play a critical role(s) in heart development/function.
Helfman et al. [12] using GFP or other tagged non-muscle TM isoforms (TM-1,-2,-3,-4,-5(NM1),-5a or -5b) and striated muscle (skeletal muscle α-TM) isoforms have shown that these non-muscle TM isoforms are incorporated into actin filaments in neonatal rat cardiomyocytes (NRC). All of these non-muscle TM isoforms, were localized into the I-band of NRCs. In other word, these researchers showed that specific isoforms characteristic of nonmuscle cells can incorporate into the myofibrillar apparatus of cardiomyocytes. Incorporation of these GFP.nonmuscle TM fusion proteins into the myofibers exhibited sarcomeric shortening and cell beating. However, they did not address whether the nonmuscle isoforms can substitute functionally for sarcomeric TM proteins or not. Following their lead, we are planning to study the myofibrillogenesis in axolotl hearts with GFP. TPM2η or other novel TPM2 isoforms. Mexican axolotl (Ambystoma mexicanum) is a unique animal model for studying structural/ functional relationships of various myofibrillar proteins especially tropomyosin. Some of these animals carry a genetic mutation in gene “c” where “c” stands for cardiac lethal. The homozygous (c/c) mutant axolotl embryos form hearts that do not beat. As a result, the embryos die after hatching due to a lack of circulation [22-25]. The mutant hearts are deficient in sarcomeric TM protein and do not form organized myofibrils. However, the transfection of TPM1α or TPM1κ expression construct into mutant axolotl hearts allows the ectopic expression of TM protein and thereby helps to form myofibrils and let the mutant hearts beat in situ. We will also perform the transfection assays in normal axolotl hearts, which will allow us to understand whether the transfected novel TM isoforms can incorporate into organized myofibrils with help of endogenous saromeric TM proteins [12,26]. Transfection assays with mutant axolotl hearts will allow us to study whether an ectopically expressed non-muscle TM protein alone can form the myofibrils as was observed with sarcomeric tropomyosin or an ectopic overexpression of non-muscle TM disarrays the organized myofibrils that subsequently affect the contractility of the normal axolotl hearts in situ.
It is worth mentioning at this point that Assinder et al (2010) reported the expression of a novel TPM2 splice variant (identical with our TPM2ε) in human prostate cancer tissues, which is absent from the normal human prostate epithelial primary cells. By Western blot analyses using TM311 antibodies they also reported the expression of a ~25kDa TM protein in human prostate tissue extracts [27]. Interestingly, TPM2ε isoform encodes a ~25 kDa protein due to the presence of a premature stop codon immediately after codon 6a. Most importantly, the ~25 kDa protein is not present in normal prostate tissues. At this juncture, we would like to point out that in a separate study, we have also detected the expression of TPM2ε transcript in three different human breast cancer cell lines but not in normal breast cell line (MCF10) (unpublished). In addition, we have detected the expression of TPM2η in one breast cancer cell line but not in normal human breast cell line (unpublished results). Hence, one cannot rule out the possibility of the association of development of cancer cell phenotype in humans.
Acknowledgement
The work was supported by a grant from the Barbara Kopp Cancer Research Fund to BJP.
References
- Gunning P, Neill G, Hardmen E (2008) Tropomyosin-based regulation of actin cytoskeleton in time and space. Physiology Review 88:1-35.
- Lees-Miller J, Helfman D (1991) The molecular basis for tropomyosin isoform diversity. Bioessays 13: 429-437.
- Perry, SV (2001) Vertebrate tropomyosin, properties and function. J Muscle Res Cell Motil 22: 5-49.
- Piples K, Wieczorek DF (2000) Tropomyosin 3 increases striated muscle isoform diversity. Biochemistry 39: 8291-8297.
- Pittenger MF, Kazzaz JA, Helfman DM (1994) Functional properties of nonmuscle tropomyosin isoforms. Curr Opin Cell Biol 6:96–104.
- Rajan S, Jagatheesan, Karam CN (2010) Molecular and functional 13 characterization of a novel cardiac specific human tropomyosin isoform. Circulation 121:410-418.
- Thierfelder L, Watkins H, MacRae C (1994) Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 77: 701-712.
- Tajsharghi H, Ohlsson M, Palm L, Oldfors, A (2012) Myopathies associated with btropomyosin mutations. Neuromascular Disorders 22: 923-933.
- Clarke NF, Kolski H, Dye DE et al. (2008) Mutations in TPM3 are a common cause of congenital fiber type disproportion. Ann Neurol 63: 329-337.
- Laing NG, Wilton SD Akkari PA (1995) A mutation in the Alpha-tropomyosin gene TPM3 assocated with autosomal dominant nemalin myopathy. Nat Genet 9: 75-79.
- Schevzov G, Whittaker S, Fath T, Lin J.J-C, Gunning P.W. (2011) Tropomysin isoforms and reagents. BioArchitecture. 1: 135-164.
- Helfman DM, Berthier C, Grossman J, Leu M, Ehler E et al (1999) Nonmuscle tropomyosin 4 requires coexpression with other low molecular weight isoforms for binding to thin filaments in cardiomyocytes. J Cell Science 112: 371-380.
- Denz C. R, Zhang C, Jia P, Du J, Huang X et al (2011) Absence of mutation at the 5'-upstream promoter region of the TPM4 gene from cardiac mutant axolotl (Ambystoma mexicanum). Cardiovasc Toxicol. 11:235-243.
- Luque EA, Lemanski LF, Dube DK (1994) Molecular cloning, sequencing and expression of atropomyosin form cardiac muscle of the Mexican axolotl, Ambystoma mexicanum. Biochem.Biophys.Res.Comm 203: 319-325.
- Zajdel RW, McLean MD, Dube S, Dube, DK (2013) Expression of tropomyosinin relation to myofibrillogenesis in axolotl hearts. Regenerative Medicine Research 1:8 (www.regenmedres.com/content/1/1/8)
- Cooley BC, Bergtrom G (2001) Multiple Combinations of Alternatively Spliced Exons in Rat Tropomyosin-! Gene mRNA: Evidence for 20 New Isoforms in Adult Tissues and Cultured Cells. Archives of Biochemistry and Biophysics 390: 71-77.
- Davis S, Watson J.C. (1996) In vitro activation of the interferoninduced,double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human alpha-tropomyosin. Proc. Natl Acad. Sci.USA 93:508–513.
- Jean M. Nussbaum, Shobha Gunnery, Michael B. Mathews (2002) The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the doublestranded RNA-dependent protein kinase PKR. Nucleic Acids Res 30: 1205-1212.
- Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nature Reviews Molecular Cell Biology 5: 827-834.
- Zhu SZ, Si M-L, Wu H, Mo Y-Y (2007) MicroRNA-21 Targets the Tumor Suppressor Gene Tropomyosin 1 (TPM1). J. Biol. Chem 282: 14328-14336.
- Colby SA, Bowers SLK, Troy BA (2009) Cardiac Fibroblast: The RenaissanceCell. Circ. Res 105: 1164-1176.
- Lemanski LF (1973) Morphology of developing heart in cardiac lethal mutant Mexican axolotls, Ambystoma mexicanum. Developmental Biology 33:312–333.
- Lemanski, L.F (1979) Role of tropomyosin in actin filament formation in embryonic salamander heart cells. J Cell Biol 82: 227-238.
- Lemanski LF, Nakatsugawa M, Bhatia R, Erginel-Unaltuna N, Spinner BJ et al ( 1996) A specific synthetic RNA promotes cardiac myofibrillogenesis in the Mexican axolotl.Biochem Biophys Rese Commun 229: 974–981.
- Zajdel R.W, McLean M.D, Lemanski S.L, Muthuchamy, M, Wieczorek et al (1998) Ectopic expression of tropomyosin promotes myofibrillogenesis in mutant axolotl hearts. Dev Dyn 213:412-420.
- Zajdel RW, McLean MD, Lemanski LF, Dube, DK (2000) Alteration of cardiac myofibrillogenesis by lipofectin-mediated delivery of exogenous proteins and nucleic acids into whole embryonic hearts. Anat.Embryol 210: 217-228.
- Assineder SA, Au E, Dong Q, Winnick C (2010) A novel splice variant of the b-Tropomyosin (TPM2) Gene in Prostate Cancer. Mol carcinogeness 49: 525-531.